Naive Bayes is a supervised machine learning algorithm base on Bayes Theorem.
Naive because it assumes all of the predictor variables to be completely independent of each other.
Bayes Rule:
P(A|B) = \frac{P(B|A)P(A)}{P(B}
P(A|B): Conditional probability of event A occuring, given event B
P(A): Probability of event A occuring
P(B): Probability of event B occuring
P(B|A): Conditional probability of event B occuring, given event A
In Naive Bayes, there are multiple predictor variables and more than 1 output class.
The objective of a Naive Bayes algorithm is to measure the conditional probability of an event with a feature vector
Problem Statement:
Predict employee attrition
Create training (70%) and test (30%) sets and utilize reproducibility.
Distribution of Attrition rates across train and test sets.
No Yes
0.8394942 0.1605058
No Yes
0.8371041 0.1628959 Notice that they are very similar.
A Naive Overview
The Naive Bayes classifier is founded on Bayesian probability, which incorporates the concept of conditional probability.
In our attrition data set, we are seeking the probability of an employee belonging to attrition class C_{k} given the predictor variables x_{1}, x_{2}, ... x_{n}
A posterior:
Posterior = \frac{Prior * Likelihood}{Evidence}
Assumption:
The Naive Bayes classifier assumes that the predictor variables are conditionally independent of each other, with no correlation.
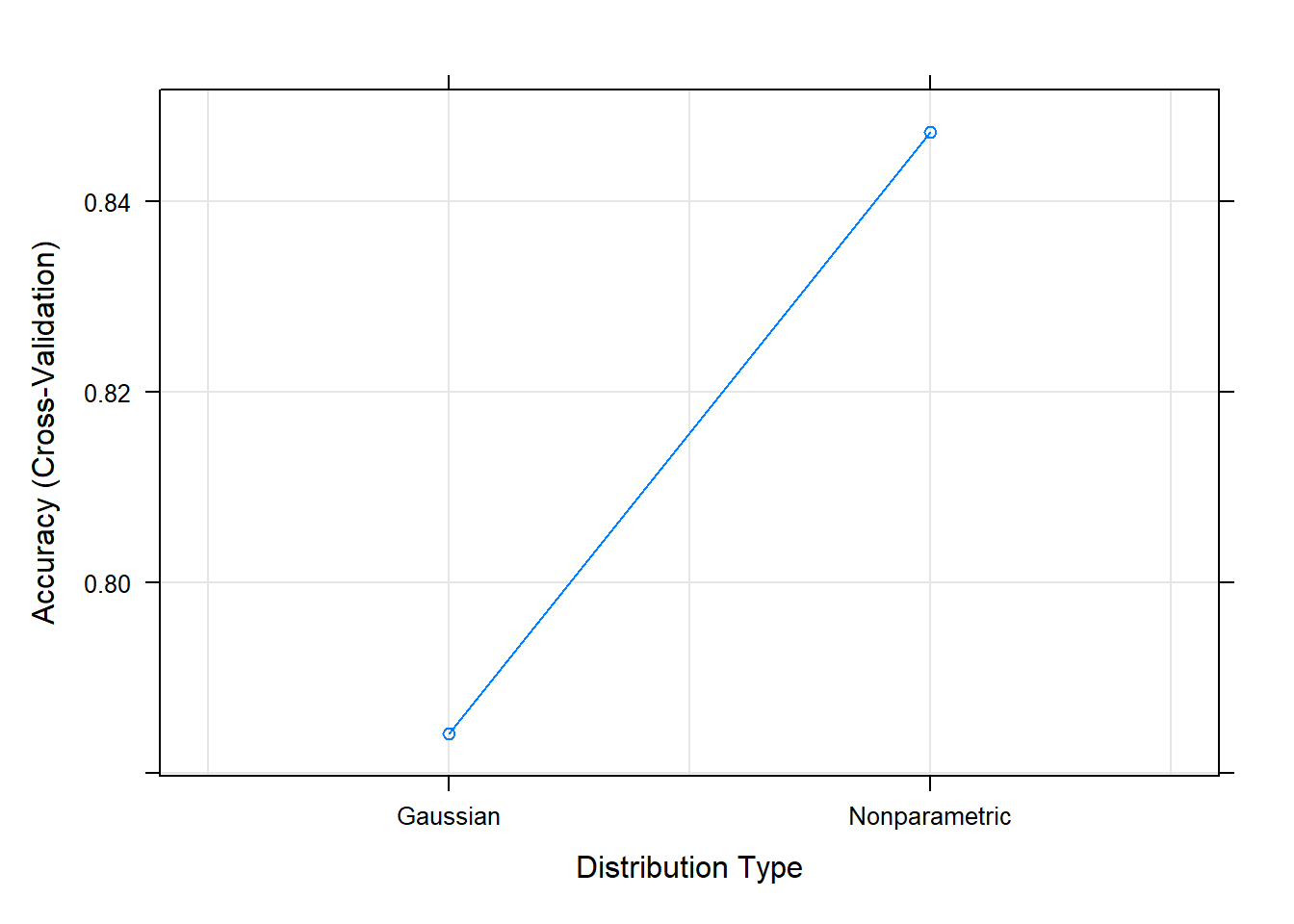
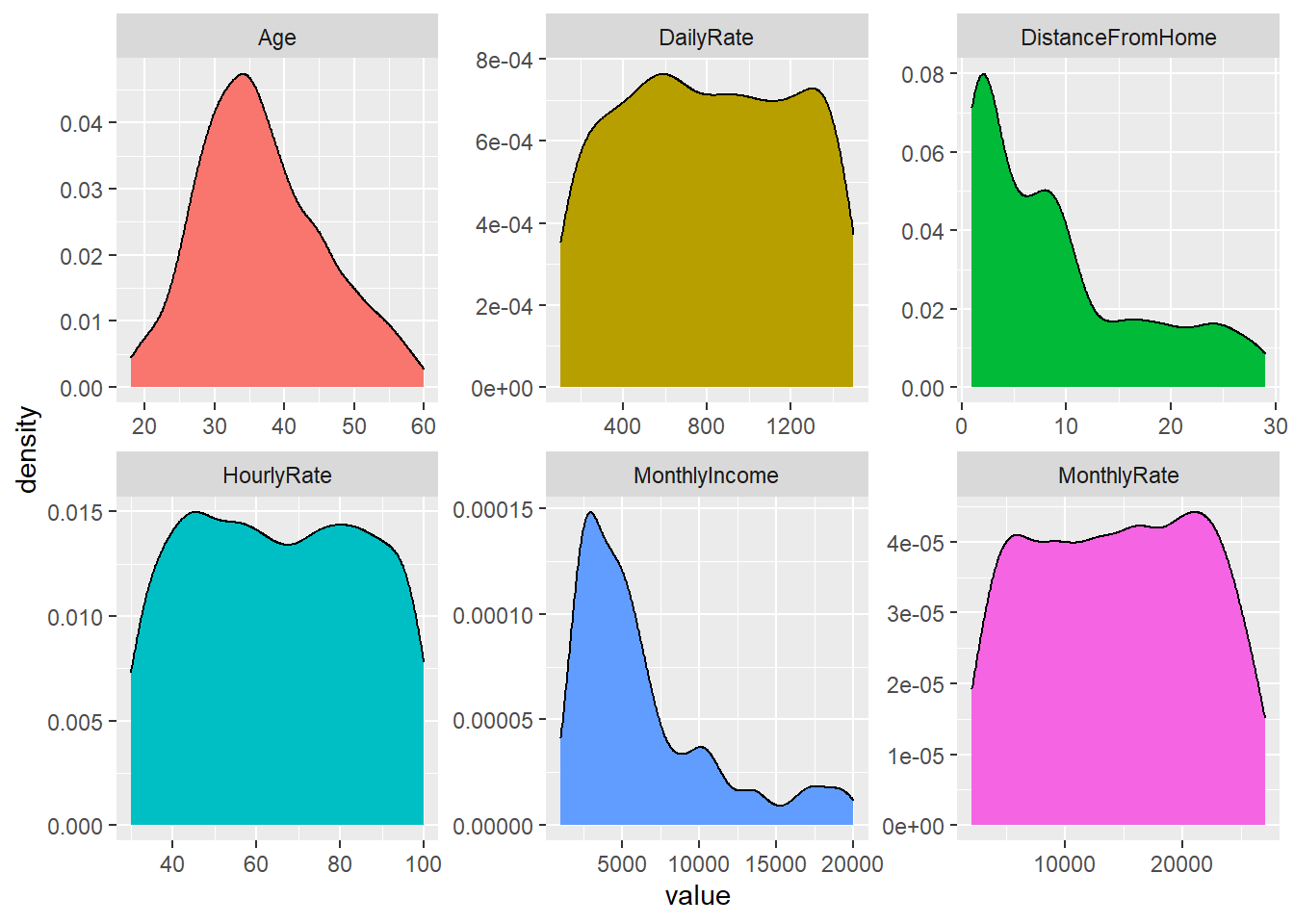
An assumption of normality is often used for continuous variables.
But you can see that normality assumption is not always held:
Code
Advantages and Shortcoming
The Naive Bayes classifier is simple, fast, and scales well to large n.
A major disadvantage is that it relies on the often-wrong assumption of equally important and independent features.
Implementation
We will utilize the caret package in R.
- Create response and feature data
Initialize 10-fold cross validation using caret’s trainControl function.
Training the Naive Bayes model:
#Confusion Matrix to analyze our results:
Cross-Validated (10 fold) Confusion Matrix
(entries are percentual average cell counts across resamples)
Reference
Prediction No Yes
No 76.8 8.2
Yes 7.1 7.9
Accuracy (average) : 0.8473To test the accuracy on our test data set:
Confusion Matrix and Statistics
Reference
Prediction No Yes
No 329 38
Yes 41 34
Accuracy : 0.8213
95% CI : (0.7823, 0.8559)
No Information Rate : 0.8371
P-Value [Acc > NIR] : 0.8333
Kappa : 0.3554
Mcnemar's Test P-Value : 0.8220
Sensitivity : 0.8892
Specificity : 0.4722
Pos Pred Value : 0.8965
Neg Pred Value : 0.4533
Prevalence : 0.8371
Detection Rate : 0.7443
Detection Prevalence : 0.8303
Balanced Accuracy : 0.6807
'Positive' Class : No